Abstract
Introduction Acute kidney injury (AKI) in hospitalized patients is associated with a high morbidity and mortality. Individuals with sickle cell disease (SCD) frequently experience organ dysfunction, including AKI, during hospitalization for episodes of vaso-occlusive crisis (VOC). AKI can lead to progressive kidney disease and chronic kidney disease (CKD), which is associated with further increased morbidity and mortality. Identifying AKI early during hospitalizations can lessen the risk of progressive CKD and renal replacement therapy. Utilizing machine learning techniques, we hypothesized that risk of AKI can be identified in SCD patients before the onset of kidney injury, leading to earlier recognition of AKI.
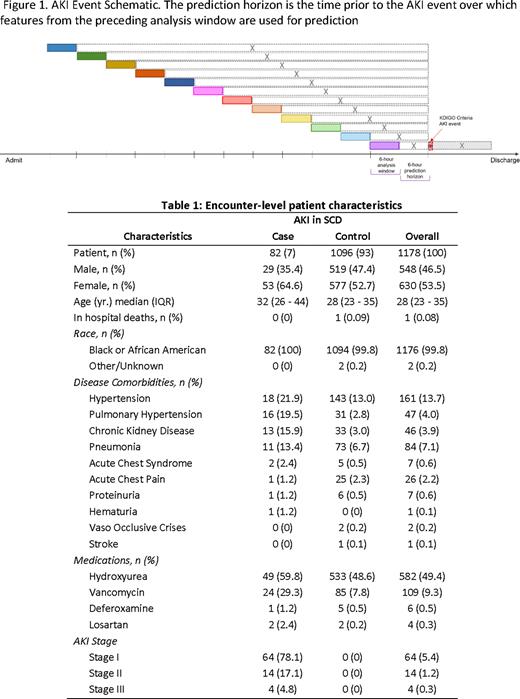
Methods This is a single-site, retrospective IRB-approved study of 6,278 patient encounters with SCD admitted across five regional hospitals in Memphis, TN, from July 2017 to December 2020. Of these, 1,178 patient encounters were selected for analysis by including encounters with continuous minute-by-minute physiological data availability and excluding dialysis-dependent patient encounters. Each patient admission was considered a separate patient encounter if the interval between admissions was at least 1 month. AKI was defined based on the 2012 Kidney Disease Improving Global Outcomes (KDIGO) clinical practice guideline. Physiomarkers, a set of signal processing features, derived from five physiological data streams from bedside monitors, including heart rate, respiratory rate, and blood pressure (systolic, diastolic, and mean), were used, along with clinical laboratory and demographic data. Missing laboratory values and vital sign measurements were first imputed using the last-one carry forward imputation. The remaining missing values were imputed by the global median of the associated variable in the training dataset. Logistic regression, support vector machine, random forest, and eXtream Gradient Boosting (XGBoost) classifiers were used for classification. Case (AKI) and control records were divided into 12 non-overlapping but consecutive 6-hour analysis time-windows (Figure 1). Prediction horizons (the time in advance each model bundle was asked to predict the AKI event) were increased by 6-hour until the beginning of the record. Controls were balanced with cases using a bootstrap method and randomized across each unique iteration to minimize selection bias. Statistical features were derived from the 6-hour analysis window immediately prior to each prediction horizon. Explainability of models generated at each time window was assessed using averaged Shapley Additive Explanations (SHAP) scores.
Results A total of 82 (7%) patient encounters were identified as AKI cases based on the 2012 KDIGO guidelines. The other 1096 SCD patient encounters served as controls (Figure 1). Overall, there were more females (64.6%) than males among AKI cases (Table 1). A history of hypertension, pulmonary hypertension, CKD, and pneumonia were higher in AKI cases than in controls (Table 1). The XGBoost model most accurately predicted AKI up to 60 hours before AKI onset, with an average test sensitivity, specificity, and area under the receiver operating characteristics curve of 0.86, 0.72, and 0.80, respectively.
Conclusions In this proof-of-concept study, we found that physio/biomarkers derived from continuous bedside monitoring and laboratory data are temporally and differentially expressed in SCD patients with AKI. A minimalistic artificial intelligence model can be developed using this information to predict AKI earlier in hospitalized SCD patients and may enable the development of innovative prevention strategies.
Disclosures
Lebensburger:Forma Therapeutics: Consultancy; Novartis: Consultancy; BPL: Consultancy; Agios Pharmaceuticals: Consultancy. Ataga:Biomarin: Consultancy; Roche: Consultancy; Agios Pharmaceuticals: Consultancy; Forma Therapeutics: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Pfizer: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal